GPUs Help Unravel “Origami-like” Behavior of Human Genome
OREANDA-NEWS. Albert Einstein. Martha Stewart. Dale Earnhardt, Jr. The guy who helps you with your taxes.
The information needed for the development and functioning of each of them – and of all living things – is in our genomes.
This genetic “instruction book” contains about 3 billion base pairs of DNA. That includes all the information needed for our bodies to function, grow, fight off diseases and do millions of other things. But while the genome in every cell of the body is identical, each type of cell needs to be different to serve its specific purpose.
So how do the genes within, say, skin cells, or lung cells or muscle cells, turn on the functions they need? How do they turn off the ones they don’t? Researchers aided by NVIDIA Tesla GPUs – part of the Tesla accelerated computing platform of GPU accelerators and enabling software – just unraveled a key piece of this mystery.
Genetic “Folding” Determines Cell Function
The Japanese word “origami” comes from the words ori, or “folding,” and kami, for “paper.” A skilled artist can turn flat piece of paper into a crane, flower, or a samurai warrior by folding it.
Turns out the human genome works in much the same way.
Using NVIDIA GPUs, researchers from Baylor College of Medicine, Rice University, MIT and Harvard University have for the first time mapped how the human genome folds in unprecedented detail.
They created the first high-resolution, 3-D maps of entire folded genomes. Among their discoveries: the genome works its biological magic with just a few basic folds—including the so-called “3D loop.”
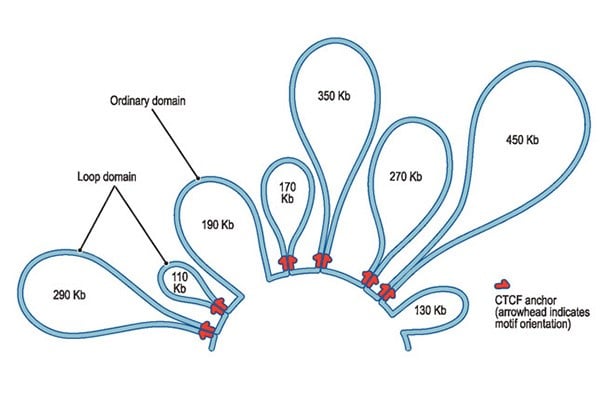
And they learned that these loops and other genome folding patterns are an essential part of genetic regulation. Just by folding the genome into different shapes, genes can be switched on or off. This lets cells take on a wide range of functions.
CPUs Not Right for the Job
To analyze the staggering amount of genomic data the research team relied on high-performance computer clusters and custom algorithms.
“We faced a real challenge because we were asking, ‘How do each of the millions of pieces of DNA in the database interact with each of the other millions of pieces?’” said Miriam Huntley, a doctoral student at Harvard’s School of Engineering and Applied Sciences. “Most of the tools that we used for this paper we had to create from scratch because the scale at which these experiments are performed is so unusual.”
The team soon realized CPUs don’t pack enough power to get the job done.
“Ordinary computer CPUs are not well-adapted for the task of loop detection,” said Suhas Rao, a researcher at Baylor’s Center for Genome Architecture. “To find the loops, we had to use GPUs, processors that are typically used for producing computer graphics.”
 Using GPUs, the researchers could study the loops and genomic folding process in a level of detail that was once impossible.
Using GPUs, the researchers could study the loops and genomic folding process in a level of detail that was once impossible.
This new information might provide new clues for cell function as well as new approaches to combat cancer and other complex diseases.
To learn more about this landmark research, check out the team’s paper in Cell Magazine (Worth noting: Rao, Huntley, and Aiden’s co-authors are Neva C. Durand, Ivan D. Bochkov, Adrian L. Sanborn, Ido Machol and Arina D. Omer, at the Baylor College of Medicine, and Elena K. Stamenova, James T. Robinson, and Eric Lander, at the Broad Institute.)




Комментарии